We show some examples below of adaptivity in an N cell network of coupled odd logistic maps f(x)=ax(1−x2).
(We should emphasize that what we show is but a small sample of some of the remarkable dynamics that can occur!)
The odd logistic map is scaled to give a self-map of
[0,1], computations are done mod 1 and the coefficient a∈[−3,3]. We consider cases where cells are identical and
approximately identical.
Coupling strengths (weights) are limited to lie in the
range [0, 2.00] and then multiplied by c/number of cells where c > 0.
Initial conditions are chosen randomly in the interval
[0.1,0.8]; initial weights are chosen randomly and lie in [0.3,0.8]. Coupling is either all-to-all or with a given
sparseness which ranges zero (no coupling) to 1 (all-to-all). When we rescale we keep c×sparseness constant.
We show dynamics of the cells and also weight averages for each node. The weight average measures the average over all inputs
to a given node. It is also possible to show the evolution of the time averages of weights (we don't show that here). Currently
we can simulate between N=4 and N=10,000 cells.
We use various types of adaption (a total of 12 currently) including 'global' schemes based on work of Ito-Kaneko, 2002, 2003 and
what we term 'fake STDP'. For the images we show here,
we strengthen weights (both ways) between cells i and j if states of i and j
are close and weaken if they are not close (we call an asymmetric directed version of this
rule fake STDP).
Typically we use adaptation of the form
wn+1ij=(1+C(1−D|xi−xj|))wnij,
where C,D>0 and xi denotes state of cell i at time n (or n+1), and wnij is the weight at
time n for the connection from cell j to cell i.
We note that the size of this computation per iteration grows like aN2, where a is independent of N (a is at least 10). This is a significant
computation for a 'desktop' computer when N = 10,000 and we do say 12,000 iterations (order 1013 computations).
Almost all the computations we show here
were done in extended precision (long double) and are sensitive to small quantities of the order 10−4900. This is necessary
to pick up some of the heteroclinic phenomena over extended time periods. The software is based on prism, used in the past for computation
of symmetric attractors and patterns, and is partly parallelized (threaded).
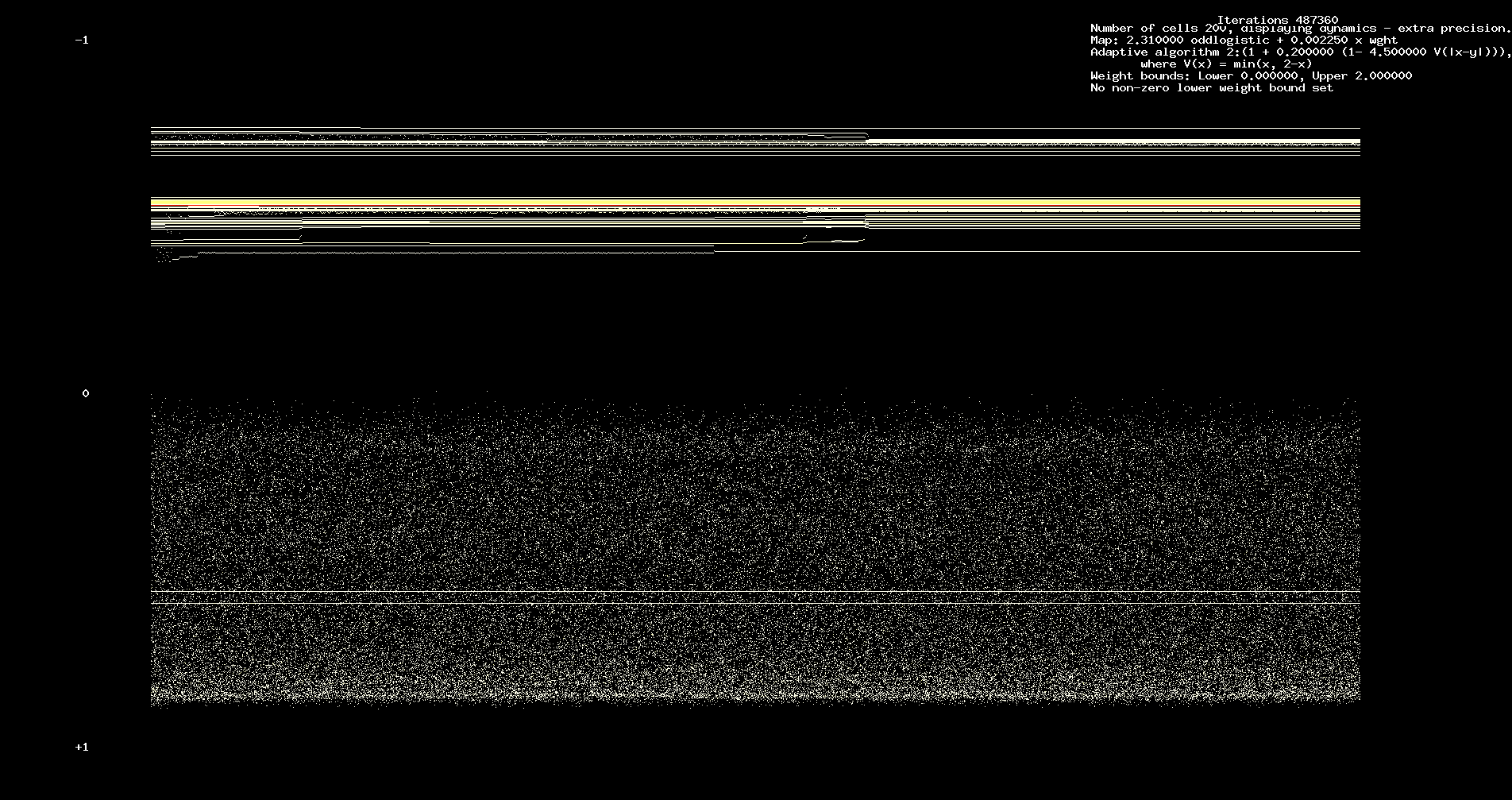
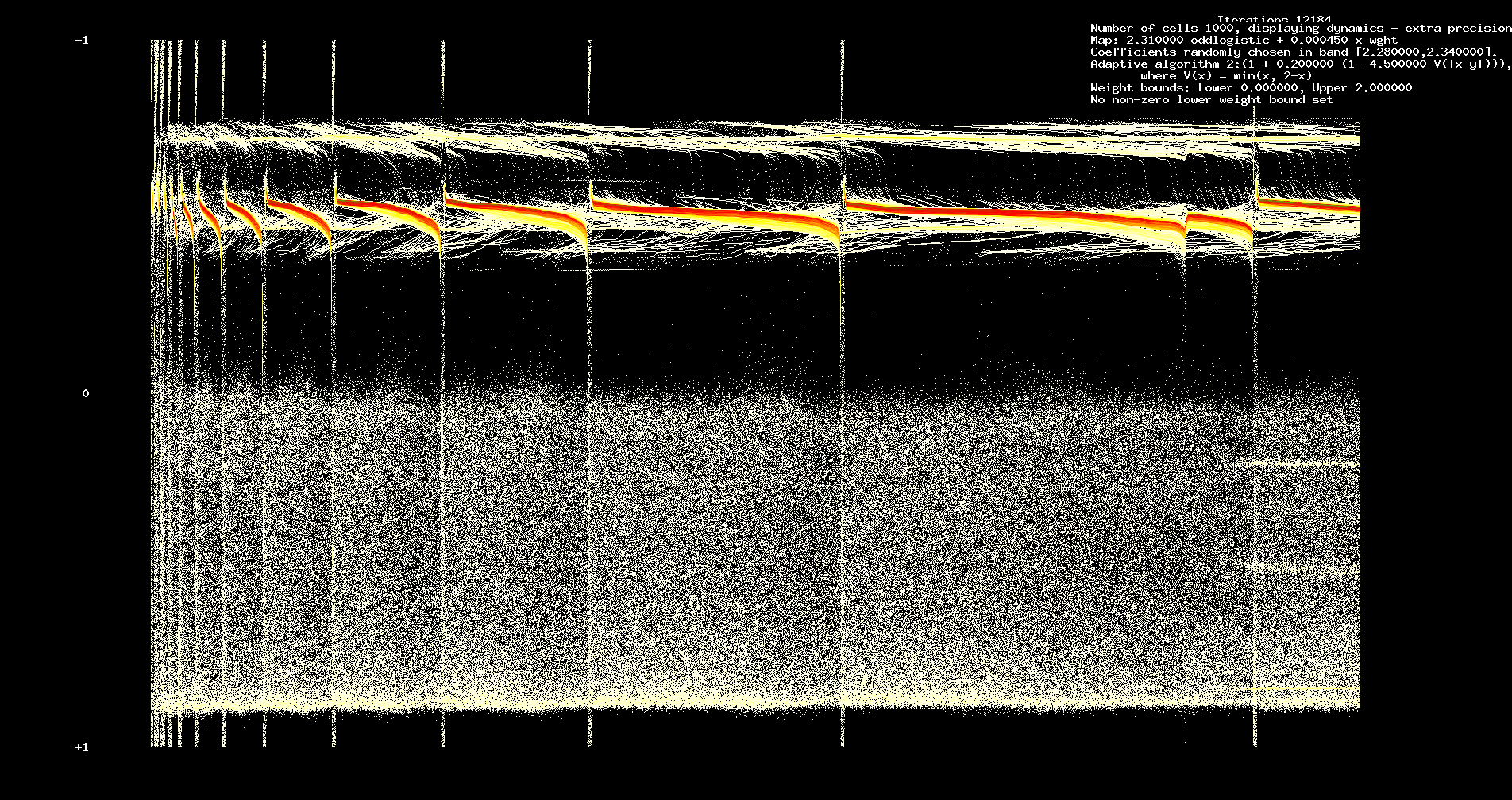
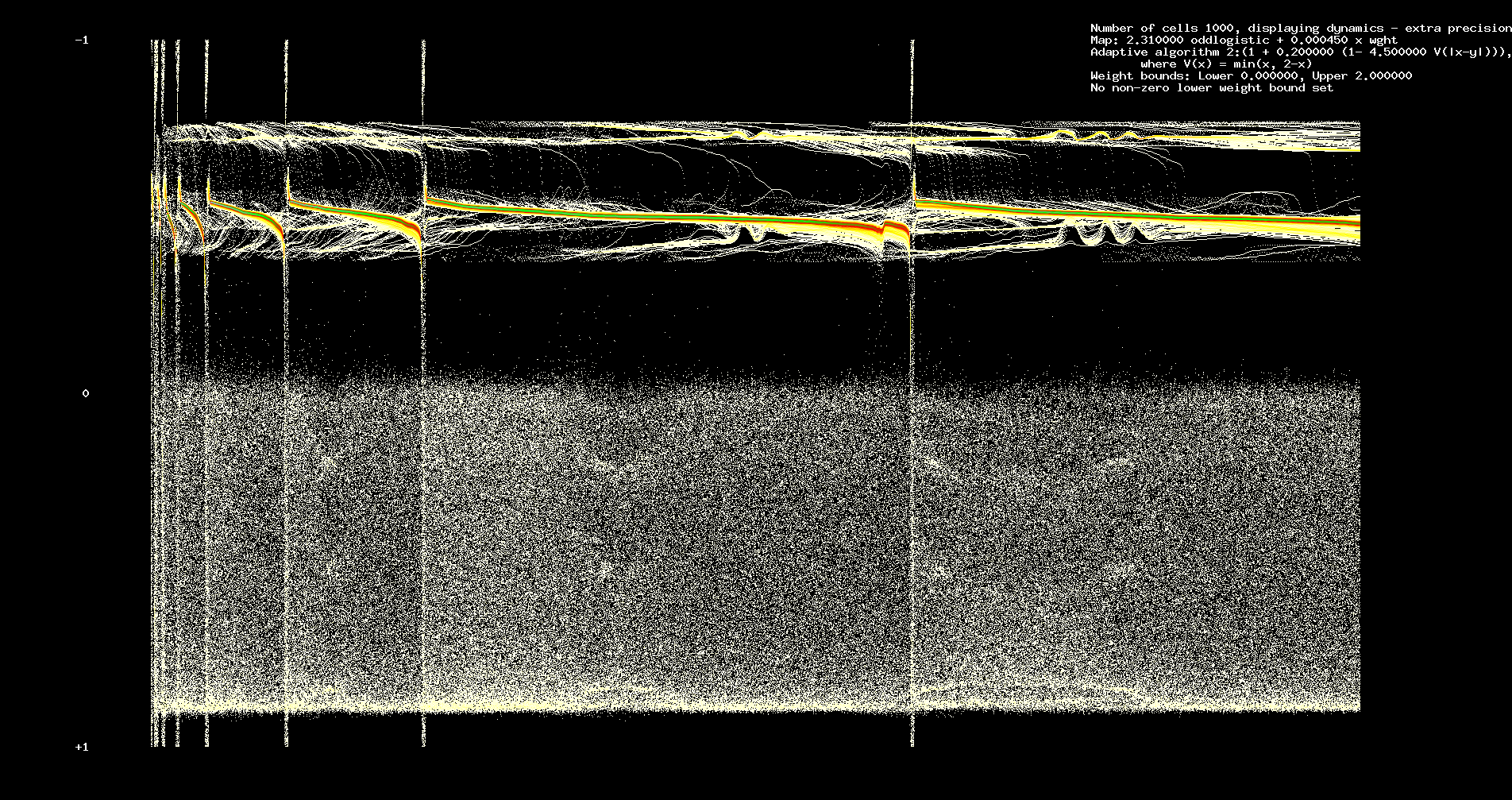
In Figures 1 and 2 we assume 200 cells, 12184 iterations. We assume identical odd logistic maps with a = 2.31.
Figure 1 shows dynamics, Figure 2 the evolution of averages of in weights from each cell.
Figure 1. Dynamics, a = 2.31
Figure 2. Evolution of average in-weights at nodes.
We remark the complexity of the initial transient which shows characteristic features of a heteroclinic cycle (between chaotic sets). However,
the heteroclinic phenomena is closely tied to the adaptivity. Run for a sufficiently long time, the network will reach a `steady state'.
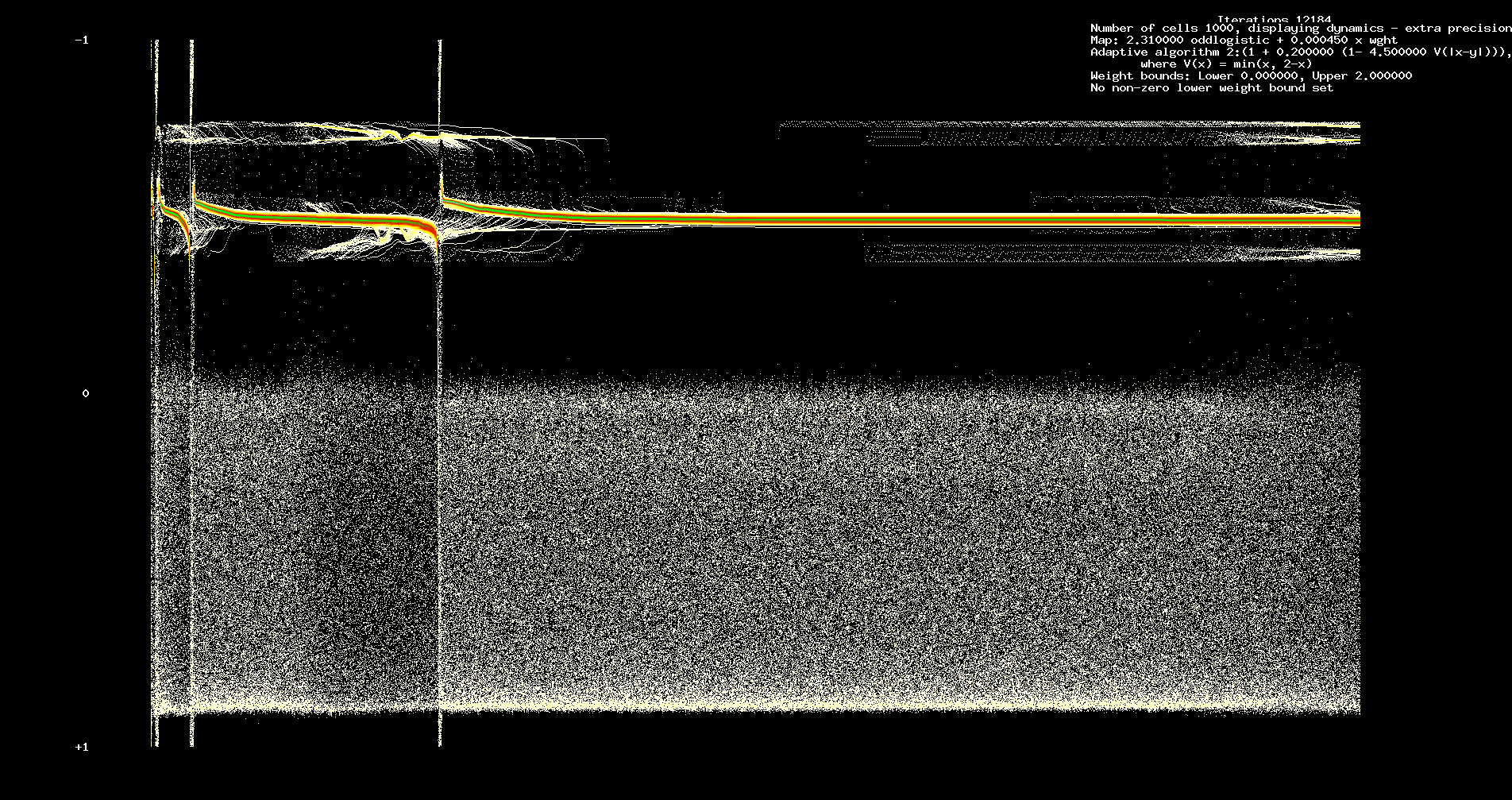
See Figure 3 below which shows dynamics over an interval of 121,840 iterations, starting at 365,520 iterations. Further iteration leaves
the picture unchanged.
Figure 3. Long term dynamics. 121,840 iterations, starting at 365,520.
The transient and long term structure appears relatively independent of the number of cells.
In the next two figures we show the first 150,000 iterations of a network with 500 cells.
Parameters are similar to those of the 200 cell network.
Figure 4. Dynamics 500 cell network: 150,000 iterations
Figure 5. Weight averages for 500 cell network: 150,000 iterations
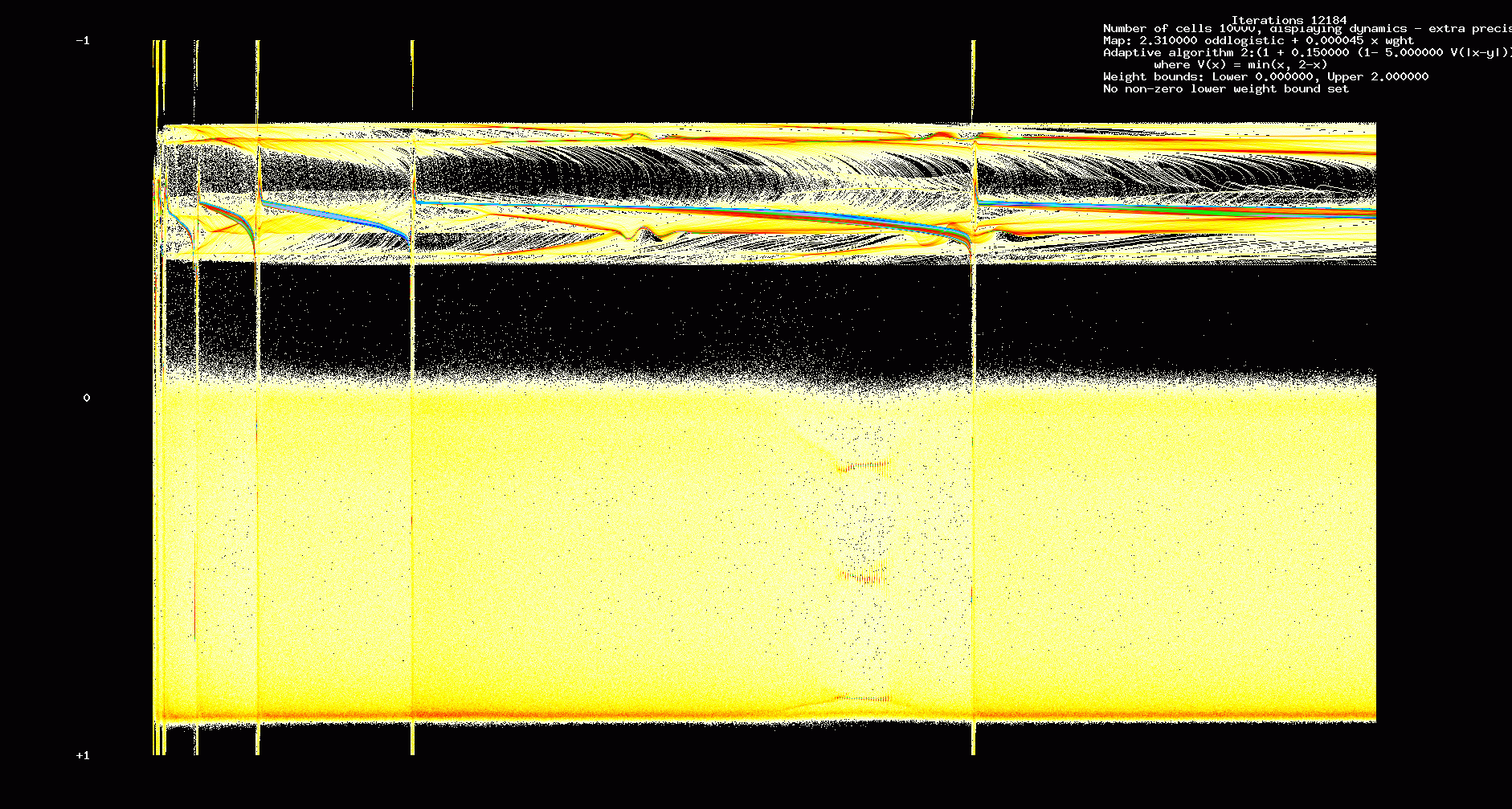
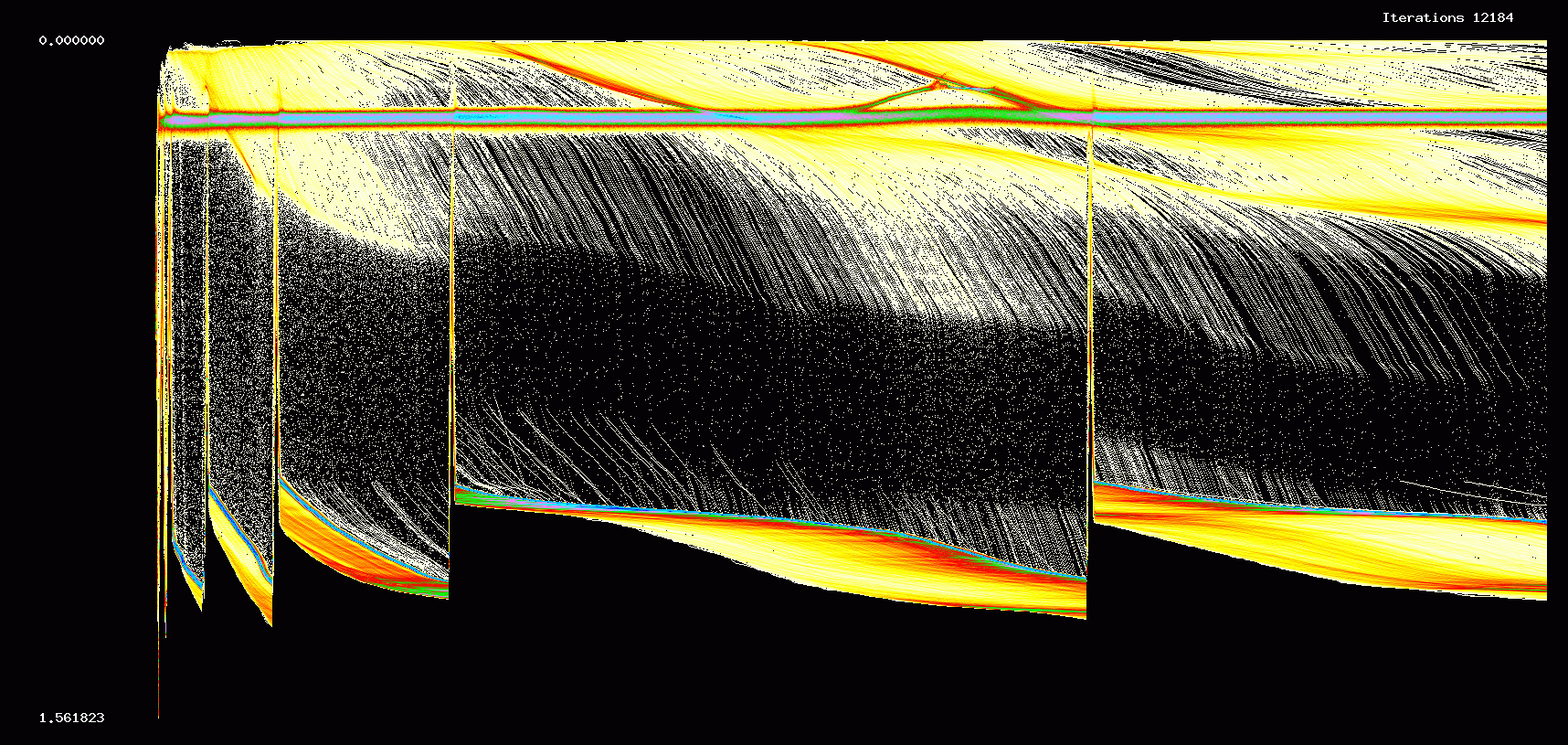
In the next two figures we show the first 12,184 iterations of a 10000 cell network with the same parameters used in Figures 1,2.
Figure 6. Dynamic transients: 10000 cell network, 12,184 iterations
Figure 7. Evolution of weights: 10000 cell network, 12,184 iterations
In the next set of figures, which are all for a 1000 cell network, we (a) break the identical cell structure and allow
for uniform distribution of the coefficients of the odd logistic map in a small interval, (b) show the effects of
decreasing the sparseness of the coupling.
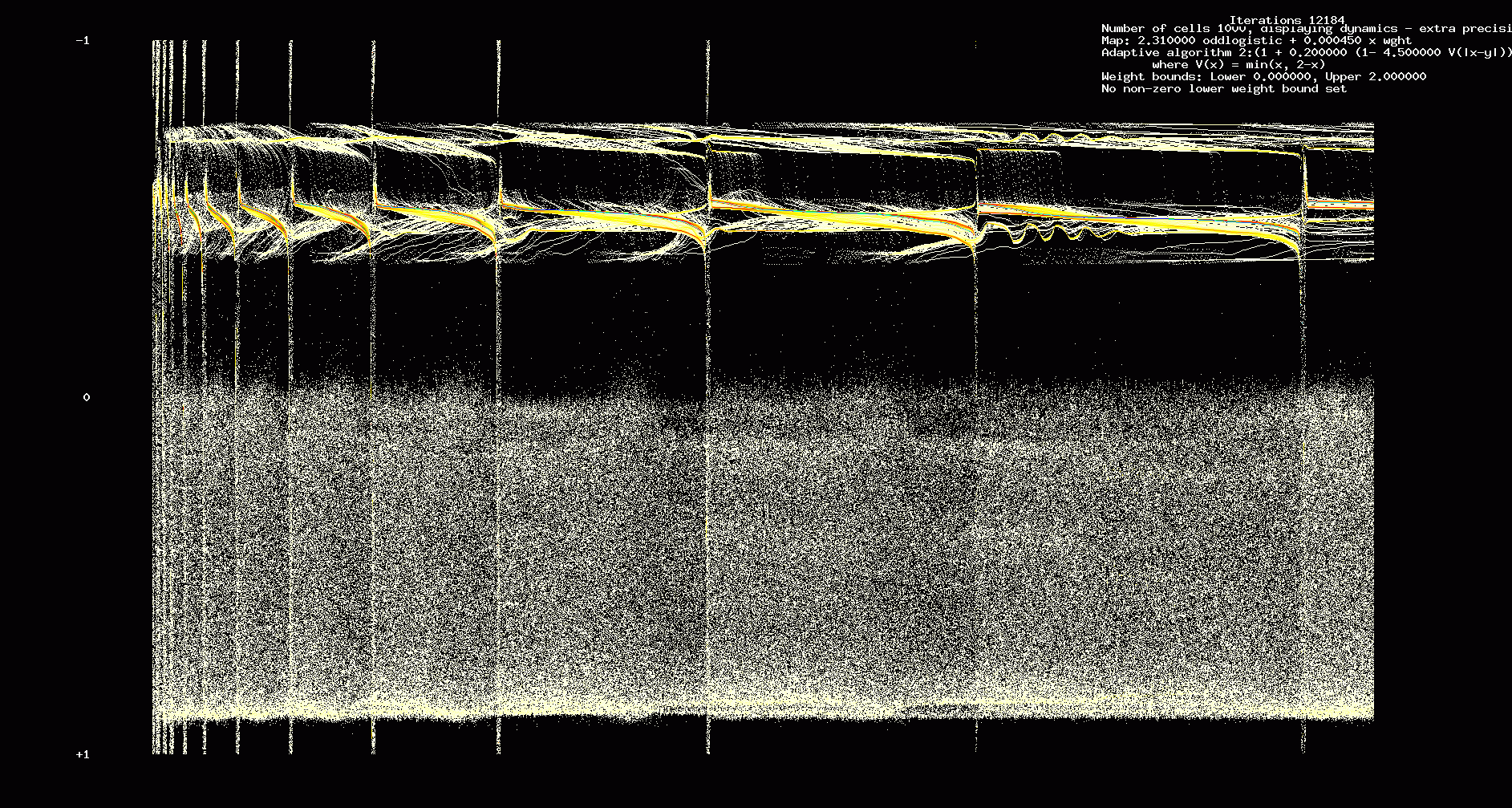
First we show the results when we assume identical odd logistic maps and all to coupling.
Figure 8. Dynamics, 1000 identical cells, sparseness = 1, 12,184 iterations
Next we show dynamics when coefficients of the logistic maps are uniformly distributed in the interval [2.28,2.34].
Figure 9. Dynamics, 1000 non-identical cells, sparseness = 1, 12,184 iterations
In Figure 10, we have identical cells but the sparseness of coupling is 0.9. As can be seen the transient behaviour is relatively unaffected.
Figure 10. Dynamics, 1000 identical cells, sparseness = 0.9, 12,184 iterations
In Figure 11, we have identical cells and the sparseness of coupling is 0.8. As can be seen the transient behaviour is now
changed.
Figure 11. Dynamics, 1000 identical cells, sparseness = 0.8, 12,184 iterations
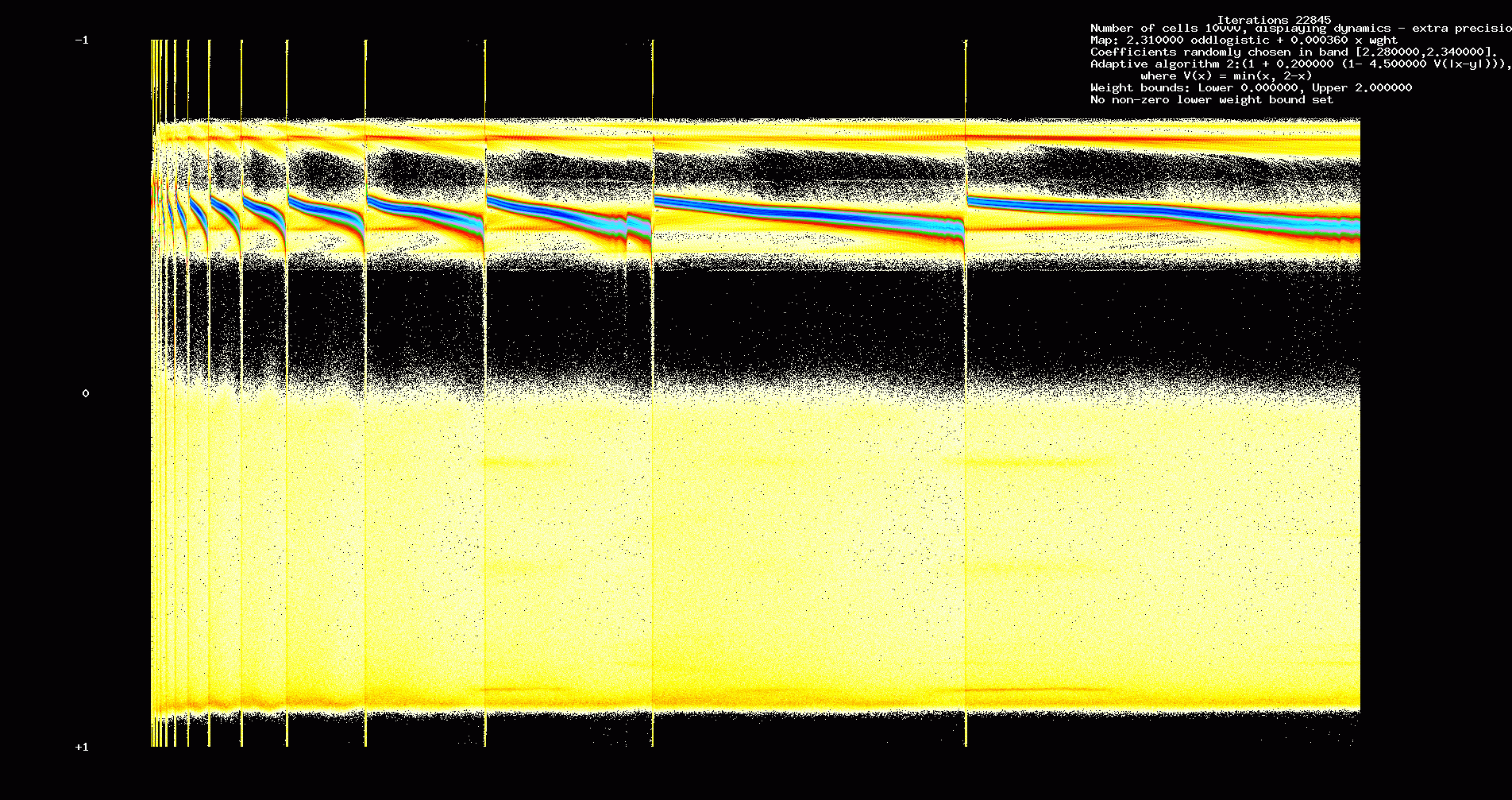
However, more can be said about this example: under a natural rescaling, heteroclinic behaviour persists even for
low sparseness and non-identical cells. In the next two
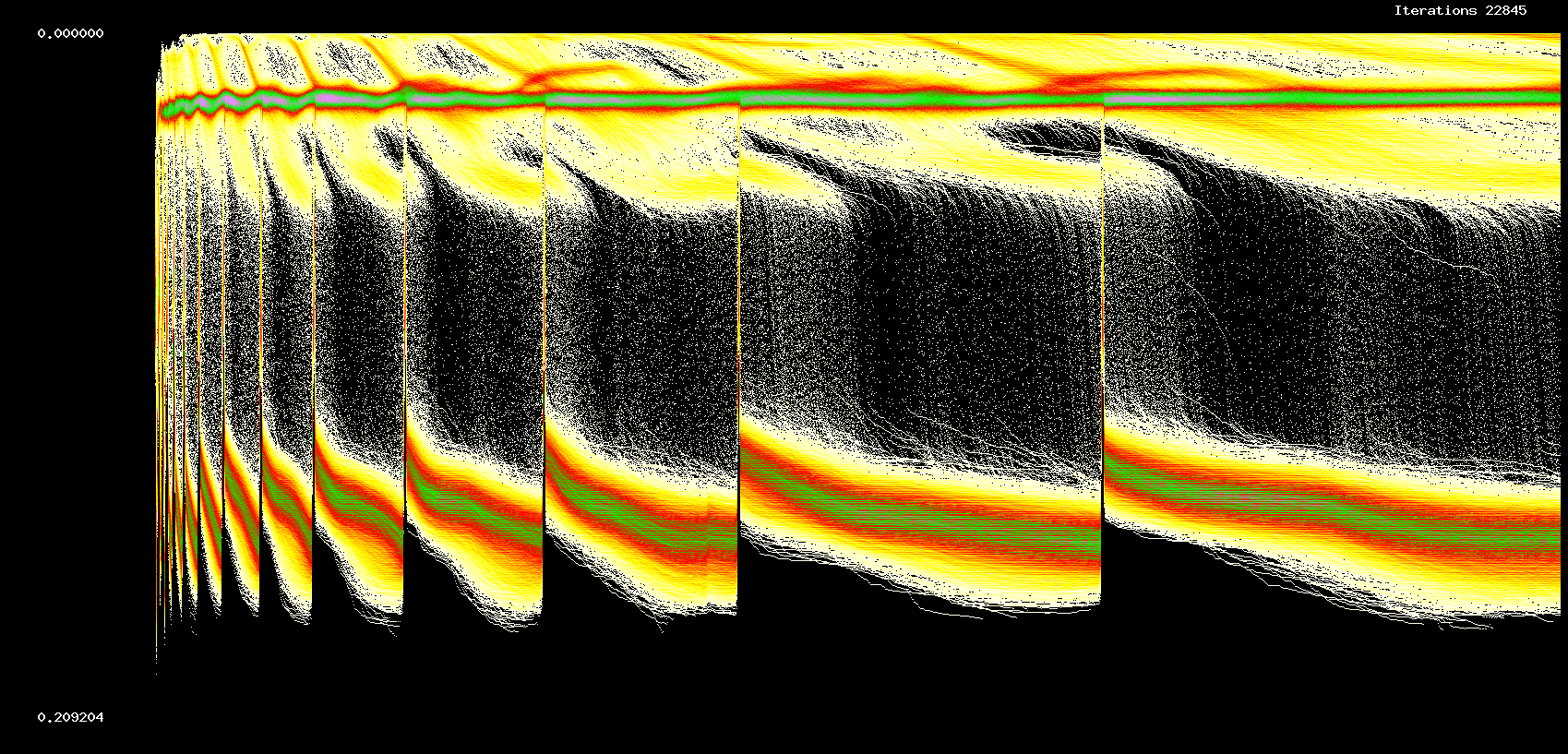
figures we show dynamics and weight evolution for a 10000 cell network, sparseness 0.125
and coefficients of the odd logistic map randomly distributed in an interval [2.31-0.03,2.31+0.03].
Fig. 12. Dynamics, 10000 non-identical cells, sparseness = 0.125, 22,845 iterations
Fig. 13. Node Weights, 10000 non-identical cells, sparseness = 0.125, 22,845 iterations
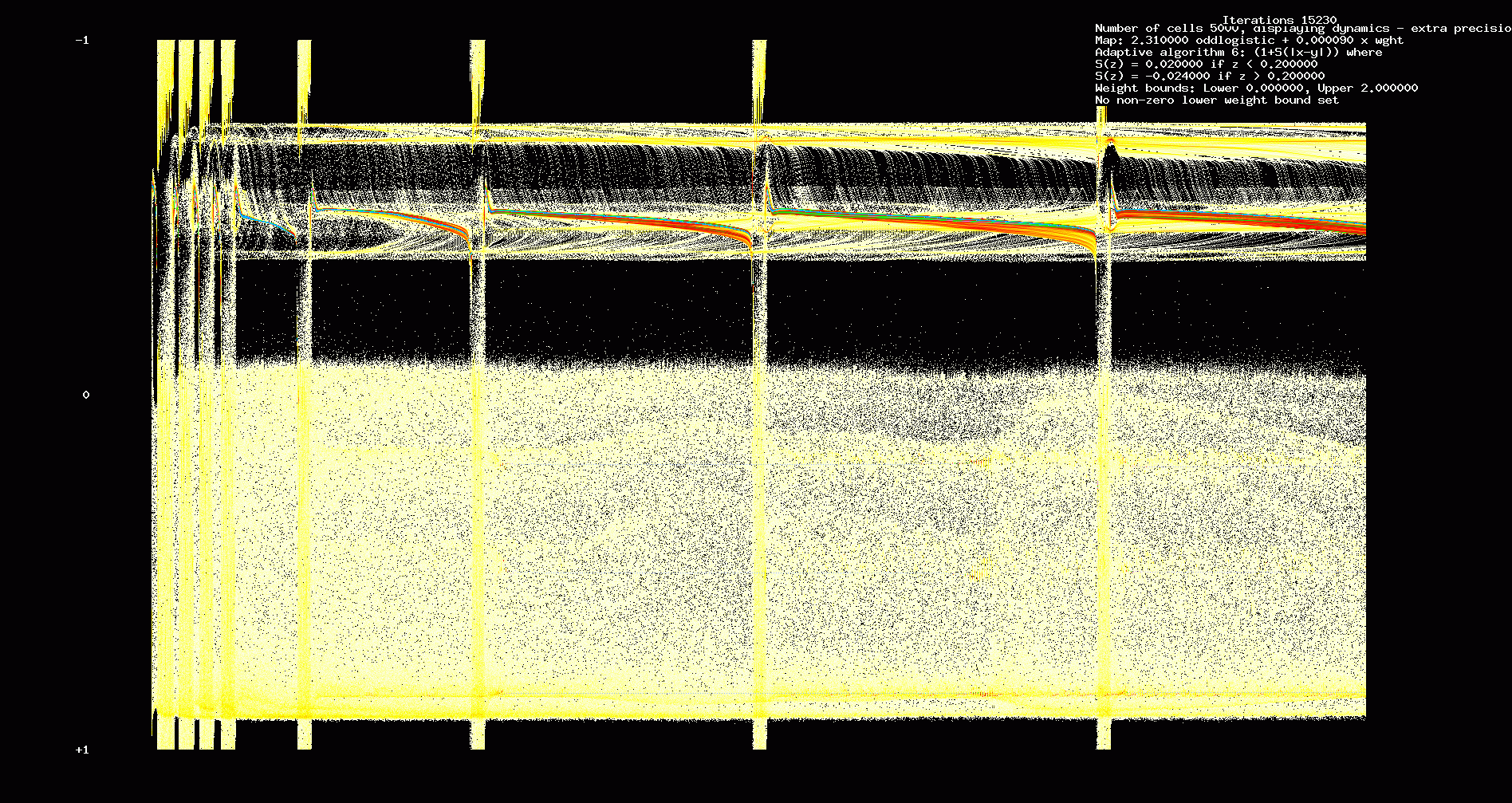
In the next two figures, we show network adaptation using a different adaptive scheme (bang-bang type control). We assume that weights evolve according to
wn+1ij=(1+W(θi,θj))wnij,
where W(x,y)=a, if |x−y|<α, W(x,y)=−b if |x−y|≥α, where
0<a<b<1 and typically a,b are small. We continue to assume that weights are constrained to lie in [0,2].
Fig. 14. Dynamics. 5000 identical cells, 'bang-bang' type adaptation, 15,230 iterations.
Fig. 15. Weights. 5000 identical cells, 'bang-bang' type adaptation, 15,230 iterations.
Next we show a simulation of a 1000 cell network over 2.534 million times steps
(points are plotted every 2000 time steps).
This is quite a big computation but the
evolution is similar to what we have shown before: eventually the system settles down to a steady state.
We use the same adaptive algorithm as in Figure 14 and assume sparseness of coupling is 0.5.
Other parameters are the same as those used for the network of Figure 14. In particular, we assume
a=2.31.
Figure 16. Dynamics, 1000 cells, 2.534 million iterations
Figure 17. Evolution of weights, summed over each node.
Synchronous and asynchronous computation and SLOGALS
A digital computer computes synchronously: computations are timed to a clock. If the computation is
threaded (or parallelized), then pieces of the computation are done synchronously but the synchronous blocks
are handled asynchonously. Globally Asynchronous Locally Synchronous (GALS for short). A classic problem with
threaded programming is getting global synchronization of the threads correct. For example, if thread A is using
computations of thread B, we need to be sure that thread B has finished the needed computations before
thread A uses them. Harder than it looks and the resulting code can be hard to debug. In general, getting
the asynchronous logic correct is much more costly than the synchronous logic. After making some mistakes,
we deliberately decided to mess up the global side by only synchronizing threads every hundred or so iterations (normally
there are 4 or 5 synchronizations per iteration.)
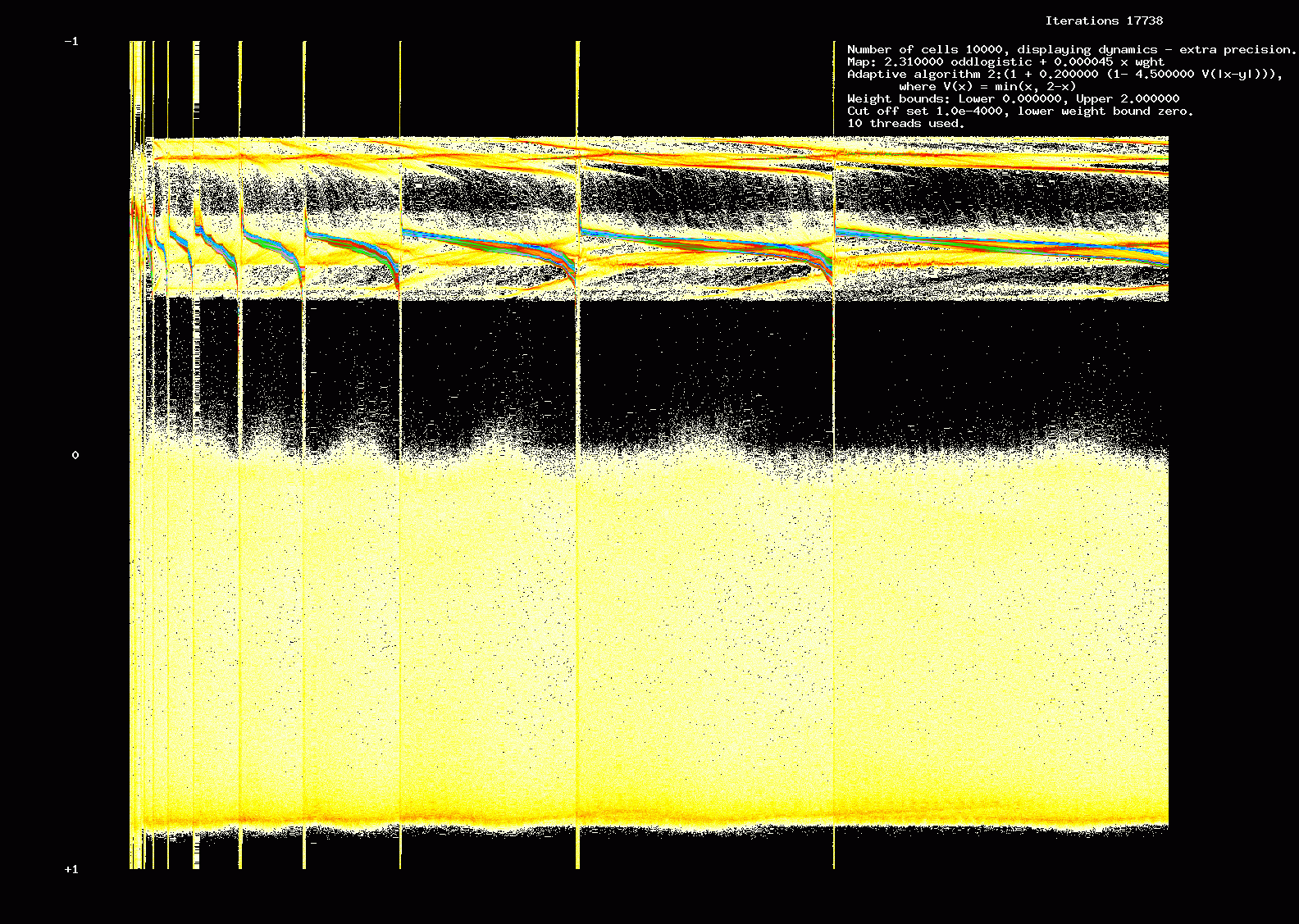
We call this a SLOGALS architecture (SLOppy GALS). We show the results of one such computation of 10,000 cells,
10 threads with synchronization every 200 iterations. The parameters are otherwise the same as for the simulation shown
in Figure 6. Moral: providing dynamics is of right sort, and we are interested in qualitative results, we can sometimes
be sloppy about the asynchronous part of the program. There is a cost: sometimes the computation will give the wrong result but
it will always complete.
Figure 18. Dynamics: 17,738 iterations, 10,000 cells SLOGALS architecture.
Research supported by NSF Grants DMS-0600927 and DMS-0806321. Software using net programs and including some components based on ATLAS and CBLAS
libraries as well as custom extra precision software.
email: mikefield@gmail.com
Professor Mike Field
Department of Mechanical Engineering
University of California
Santa Barbara, CA 93106
|